HLA molecules are the human MHC molecules. HLA molecules play a pivotal role in the normal immune response, and inability to express these molecules causes a lethal immunodeficiency. An individual’s HLA type determines in part their ability to respond to infection and predisposition to autoimmune disease. Matching donor and recipient HLA types as closely as possible improves outcome in most types of transplantation.
HLA MOLECULES – STRUCTURE AND FUNCTION HLA molecules are divided into two main classes, HLA Class I (including HLA-A, B and C) and HLA Class II molecules (including HLA-DR, DP and DQ). HLA Class I molecules have one variable a chain, and are stabilised by B2-microglobulin.
HLA Class II molecules are heterodimers, comprising 2 variable chains (a and β chains). Both Class I and Class II molecules have a peptide-binding groove (Figure 3.5). The physiological function of both classes of HLA molecules is to present short pathogen-derived peptides to T cells. This is the key interaction in initiating an antigen specific, adaptive immune response.
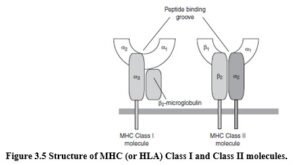
HLA molecules bind many different peptides stably as an integral part of the HLA molecule – HLA is unstable when peptides are not bound. The peptide binding domains of HLA Class I, and HLA Class II molecules are polymorphic; that is, particular amino acids in the peptide-binding grooves vary from allele to allele. These polymorphic residues make contact with the antigen-derived peptide. The peptide amino acids that bind HLA are called anchor residues. Peptides range in length from 8 to 10 residues for HLA Class I, to 12-20 residues for HLA Class II.
HLA Class I molecules are expressed on all nucleated cells and on platelets and present cytoplasmic peptides to CD8+ cytotoxic T cells. Widespread expression of HLA Class I molecules allows all cells to present peptides from intracellular pathogens to cytotoxic T cells of the immune system.
HLA Class II molecules are expressed on a restricted repertoire of immune cells including dendritic cells, monocytes and macrophages, B cells, activated T cells and thymic epithelial cells. HLA Class II molecules present antigen to CD4+ helper T cells to initiate adaptive immune responses, a role requiring restricted expression. At sites of inflammation, particularly in the presence of IFN-γ, Class II molecules are induced on cells that do not normally express these molecules.
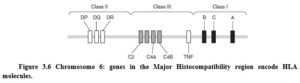
The genes encoding the HLA molecules are found in a small cluster on Chromosome 6 (Figure 3.6). This region contains in excess of 200 genes including both HLA, non-HLA genes involved in the immune response, and many genes with no apparent immunological role.
HLA MOLECULES – DIVERSITY Within the population, there are hundreds of different HLA alleles. Most variability between different alleles of a HLA molecule resides around the peptide-binding groove. Expression of HLA molecules is polygenic (we each have several HLA genes) and polymorphic (at each HLA gene locus there are many possible alleles). Individuals of differing HLA type infected with the same organism will usually present different pathogen-derived peptides to initiate an immune response. The outcome of the immune response is often equivalent, however, possession of particular HLA types has been associated with disease resistance in some situations. For example, expression of HLA-B53 is associated with reduced mortality from severe malaria.
We inherit one set of HLA genes from each parent, and both maternal and paternal alleles are expressed on all cells. Within particular populations gene ‘packages’ known as haplotypes are frequently identified. Thus some HLA genes may be found almost exclusively associated with typical haplotypes within a particular population. These haplotypes include HLA genes and also the intervening genes encoded on chromosome 6, many of which have immunological functions. Haplotype associations of HLA molecules differ in different populations.
Genetic diversity of HLA molecules helps to ensure that some members of a population can mount an immune response to any organism. While some individuals may be unable to eliminate a pathogen, diversity makes survival at population levels more likely. At an individual level, people who are heterozygous (expressing different maternal and paternal HLA alleles) can bind and present a greater repertoire of peptides to their T cells. It appears that progression to acquired immunodeficiency syndrome (AIDS) is slower in human immunodeficiency virus (HlV)-infected individuals who are heterozygous for HLA molecules, suggesting a possible survival advantage attributable to HLA diversity.
DISEASE ASSOCIATIONS Particular HLA types have been associated with both disease susceptibility and resistance. Associations are relative rather than absolute, and therefore are rarely useful in establishing a diagnosis in individual patients. Apparent associations may reflect a true role of the HLA molecule or may be due to linkage between the HLA allele and a physically close disease related (non- HLA) gene. In the non-immunological sleep disorder narcolepsy, an apparent HLA association was explained by linkage disequilibrium between a mutated gene encoding a hypocretin receptor and particular HLA alleles (Table 3.5).

Several non-HLA genes are encoded in the HLA region of chromosome 6, many of which affect the immune response. For example, the TNF-a gene is encoded in this region, and a functional polymorphism in this gene determines whether large or small amounts of TNF-a are produced. The high-producing TNF- a genotype is linked to HLA-DR3 and included in a haplotype associated with increased risk of autoimmunity. Determining whether an apparent HLA association is significant is complicated by the common occurrence of haplotypes. Demonstrating a true HLA effect usually requires the study of several populations where individual HLA molecules are found with different haplotype associations.




