The protective function of T cells depends on their ability to detect cells that harbour or internalise pathogens. T cells do this by responding to pathogen-derived peptides bound to MHC molecules on cell surfaces. Antigens may be endogenous (derived from molecules inside the cell, including molecules derived from intracellular pathogens) or exogenous (occurring outside the cell, but taken up by the processing cell). The generation of peptides from antigens is known as antigen processing. Not surprisingly, different mechanisms are involved in processing endogenous and exogenous antigens. The display of these peptides on the cell surface is known as antigen presentation.
ANTIGEN PRESENTING CELLS Pathogens and other exogenous antigens are internalised into the vesicular compartments of antigen presenting cells (APCs), by phagocytosis, endocytosis or macropinocytosis. APCs include professional APCs such as dendritic cells that specialise in stimulating a T cell response; non-professional APCs such as macrophages that can present antigen but have other primary functions and B cells that internalise antigen by receptor-mediated endocytosis of cognate antigen bound to their surface immunoglobulin (sIg).
MHC CLASS I AND CLASS II MOLECULES DELIVER PEPTIDES TO THE CELL SURFACE FROM DIFFERENT CELLULAR COMPARTMENTS WHERE THEY ARE RECOGNISED BY TWO DIFFERENT TYPES OF T CELL
MHC Class I molecules deliver peptides produced in the cytosol to the cell surface where they are recognised by CD8 cytotoxic T cells (TCs). In this way cytotoxic T cells kill cells infected with viruses or bacteria residing in the cytosol.
MHC Class II molecules deliver peptides from the vesicular system to the cell surface where they are recognised by CD4 helper T cells. In this way, helper T cells detect pathogens and their products taken up from the extracellular environment into the vesicular compartment of cells and activate other cells to eliminate the pathogen.
ENDOGENOUS ANTIGEN PATHWAY – MHC CLASS I Generally pathogen-derived peptides that bind MHC Class I molecules are derived from viruses that have infected host cells. Viruses take over the cell’s biosynthetic mechanisms to make their own proteins, which are degraded and synthesised as part of normal cell protein turnover.
♦Cytosolic protein degradation takes place in a multicatalytic protease complex called the proteosome (Figure 3.7).
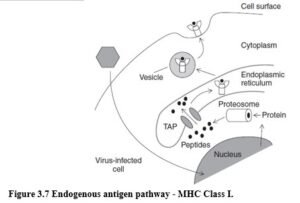
♦Degraded proteins are transported from the cytosol of a cell into the lumen of the endoplasmic reticulum (ER) via membrane transporter proteins (transporters associated with antigen processing or TAP-1 and TAP-2).
♦In this compartment peptides bind to MHC Class I molecules. Peptide binding is an essential step in the assembly of MHC Class I molecules and this process involves a number of accessory proteins with chaperone-like functions.
♦Peptide:MHC Class I complex is then transported to the cell surface where it can be recognised by cytotoxic T cells.
Some viruses have evolved ways to evade recognition by down-regulating the appearance of peptide:MHC complexes on the cell surface.
EXOGENOUS ANTIGEN PATHWAY – MHC CLASS II Extracellular pathogens and other extracellular antigens that are internalised into endocytic vesicles (endosomes) of APCs are processed in the exogenous antigen pathway (Figure 3.8). The contents of endocytic vesicles are not accessible to cytosolic proteosomes.

♦Protein degradation takes place in these vesicles which contain proteases.
♦MHC Class II molecules are prevented from binding peptides in the ER by associating with the invariant chain (Ii), which also targets the molecule to endosomes. MHC Class II molecules reach the endocytic vesicles of macrophages, B cells and dendritic cells.
♦When these endocytic vesicles fuse with peptide containing vesicles, the MHC molecules are loaded with peptides and transported to the cell surface where the peptide:MHC Class II complex can be recognised by helper T cells.
Initiation of an immune response requires activation of helper T cells by APCs with peptide bound to MHC Class II. APCs constantly take up antigen from their environment, and thus bacterial and viral antigens reach the endosomal compartment, even though this is not their normal site of infection. Additionally, several pathogens including mycobacteria and Leishmania replicate in cellular vesicles of macrophages.
RECOGNITION MOLECULES OF THE INNATE IMMUNE RESPONSE The innate immune
response recognises microbes by the presence of molecular motifs, which are not present in mammalian cells, using pattern recognition molecules (PRMs). PRMs are constitutively expressed and allow the innate immune response to be constantly effective, as there is no delay required to rearrange microbe specific receptors. Important PRMs are outlined below. A summary of molecular targets of PRMs involved in the innate immune response was presented in Table 1.8.2.
Mannose receptor The mannose receptor binds mannose in microbes and induces phagocytosis. Peptides from the microbe are processed and presented by macrophages, and can activate some T cell subtypes.
Toll receptors Toll-like receptors (TLRs) are a family of closely related proteins that recognise several microbial motifs including peptidoglycan, teichoic acids (found on gram-positive bacteria), LPS (gram-negative bacteria), arabinomannans and glucans. Binding of ligand to TLRs induces expression of immunostimulatory molecules and cytokines important in initiation of the adaptive immune response.
CD14 CD14, found on macrophages, binds LPS, a unique bacterial surface structure found in cell walls of gram-negative bacteria, together with an adaptor protein. When LPS binds CD14, macrophages are activated facilitating destruction of the microbe and secretion of cytokines, triggering a wide variety of immune responses.
Scavenger receptors These receptors are expressed by macrophages and recognise carbohydrates or lipids in bacterial and yeast cell walls.




